基于核酸分子学方法的肉类成分鉴别技术研究进展
王金斌1,2,3,李 文1,3,*,白 蓝1,3,刘 华1,3,蒋 玮1,3,吴 潇1,3,王荣谈4,唐雪明1,2,3,*
(1.上海市农业科学院,上海 201106;2.上海海洋大学食品学院,上海 200090;3.上海市农业遗传育种重点实验室,上海 201106;4.上海瑞丰农业科技有限公司,上海 201106)
摘要:近年来,肉类掺假问题频繁发生。基于核酸的分子生物学肉类成分鉴别技术已成为研究热点,其具有灵敏度高、特异性强、检测时间短以及成本低的优点。本文综述了基于核酸分子学的肉类成分种属鉴别技术在肉类掺假检验中的应用,着重于量化各种方法的检测限,并重点对实时荧光定量聚合酶链式反应(polymerase chain reaction,PCR)和数字PCR技术在动物成分鉴别定量分析的研究现状与前景做介绍。探讨不同来源的靶基因(核DNA和线粒体DNA)在动物成分鉴别中,定性和定量检测灵敏度与特异性的区别。
关键词:掺假;肉类制品;数字PCR;物种测定
王金斌, 李文, 白蓝, 等. 基于核酸分子学方法的肉类成分鉴别技术研究进展[J]. 食品科学, 2017, 38(11): 318-327. DOI:10.7506/spkx1002-6630-201711049. http://www.spkx.net.cn
WANG Jinbin, LI Wen, BAI Lan, et al. A review of current DNA-based methodologies for meat authentication[J]. Food Science, 2017, 38(11): 318-327. (in Chinese with English abstract) DOI:10.7506/spkx1002-6630-201711049. http://www.spkx.net.cn
民以食为天,食以安为先,吃放心健康绿色的肉类制品是食品质量安全的一个重要方面。食品掺假是一个老生常谈的话题,我国古代就有“挂羊头卖狗肉”的说法。为确保食品成分的真实性,质检“十二五”规划纲要明确指出,需重点加强开展食品掺假鉴别技术研究。国内市场近期曝光的多起肉类掺假事件引发了公众对食品安全的担忧。即使是在拥有世界上最严格食品安全制度的欧洲,2013年亦出现“挂牛头卖马肉”的造假现象,值得人们深思。
在肉类掺假形式层出不穷的情势下,对动物源性成分鉴别技术的研究逐步成为食品安全领域的研究热点。目前,使用的动物物种鉴别分析方法主要基于蛋白质和DNA分析。其中,蛋白质技术包括免疫[1]、色谱[2]和光谱[3]。但基于蛋白质的检测技术有一定的局限性,当材料进行热处理时,多数蛋白质会发生变性,不能满足检测的需要。而以核酸为基础的分析方法可以克服这些困难,因为DNA存在于所有生物的所有组织,且DNA比蛋白质的耐热性强,高温处理过的食品中仍能提取出片段化的DNA。另外,DNA比蛋白质具有更丰富的种间多态性,有利于品种鉴定[4]。目前,以检测DNA为基础的方法主要有:常规聚合酶链式反应(polymerase chain reaction,PCR)-凝胶电泳法、多重PCR-凝胶电泳法、PCR-随机扩增多态性DNA(random amplified polymorphic DNA,RAPD)分析、PCR-限制性内切酶片段长度多态性(restriction fragment length polymorphism,RFLP)分析、DNA条形码(DNA barcoding)、荧光定量PCR、微滴数字(droplet digital,dd)PCR方法等。本文将分别综述基于核酸检测的食品中肉类成分鉴别定性和定量技术的现状与问题。
1 基于核酸分子学方法的动物成分鉴别
1.1 常规PCR-凝胶电泳法
DNA片段经扩增后,通过琼脂糖凝胶电泳进行片段大小的检测,这是基于核酸分子学肉类成分鉴别方法中应用广泛且操作最简单的技术。其基本的实验思路为:根据不同物种细胞核或线粒体基因组序列中的特征位点设计物种特异性引物,利用PCR反应实现食品中目标基因片段的指数级扩增,继而通过电泳鉴别食品中可能的物种来源。目前利用常规PCR-凝胶电泳法对肉类成分检测的方法研究汇总见表1。
自1998年Tartaglia等[5]首次报道基于普通PCR方法检测饲料中的牛、羊源性成分至今,大量相关研究报道了应用PCR-凝胶电泳对食品中多种肉类成分的鉴别方法,检测限大部分在0.1%以下。其中,对未加工(生鲜肉)和加工过的(腌制或者热加工)肉类产品进行对比测试,虽然DNA高度受损,但两者PCR特异性引物的检测灵敏度差别不大。这是因为物种特异性引物的PCR技术的靶基因是短的DNA片段,具有简单性、特异性和高灵敏度的优点;主要的缺点是只有依赖针对靶序列的准确数据才能设计出相应的特异性引物。
表1 常规PCR-凝胶电泳法的汇总
Table 1 Reported PCR-gel electrophoresis methods

注:检测限中百分数均为质量分数。MT. 线粒体;SINE. 短散布重复序列;LINE. 长散布重复序列;/. 文献中没有描述。下同。
样品来源分析位点检测限(处理方式)参考文献反刍动物MT tRNA ATP6、ATP80.125%(加热)[5]牛MT tRNA ATP6、ATP80.04%(加热)[6]牛、猪、鸡和绵羊MT tRNA ATP6、ATP80.3%(加热)[7]牛MT tRNA ATP6、ATP80.125%(加热)[8]牛、猪、鸡和绵羊MT tRNA ATP6、ATP81%(加热)[9]牛、猪、鸡和绵羊MT tRNA ATP6、ATP80.01%(加热)[10]反刍动物、牛和哺乳动物MT tRNA ATP80.01%(加热)[11]猪、反刍动物和家禽MT 12S rRNA0.125%(加热)[12]牛、山羊、绵羊、反刍动物和鹿MT 12S rRNA0.05%(加热)[13]牛、山羊和绵羊MT 12S rRNA0.1%(加热)[14]猪、鸡、鹅、火鸡和鸭MT 12S rRNA0.1%(加热)[15]牛MT 12S rRNA0.1%(加热)[16]牛MT 12S rRNA、18S rRNA0.1% [17]牛MT 12S rRNA0.01 ng稀释DNA(未加工)[18]鹅和半番鸭MT 12S rRNA0.1% [19]牛、山羊、猪和鸡MT DNA/[20]牛MT DNA<1%(加热)[21]牛MT Cty b0.025%(加热)[22]山羊MT Cty b/[23]牛、山羊、猪、鸡和鸭MT Cty b、ATP0.1 g/100 g[24]牛、绵羊、山羊、猪、鸭和鸡MT D-loop区<1% DNA混合(加热)[25]牛和山羊MT D-loop区0.1% DNA混合(加热,未加工)[26]狐狸、貉子、水貂和家犬ND5/[27]牛DNA重复序列(浮力密度1.709 g/cm3)34 fg DNA(未加工);0.3 pg DNA(加热)[28]牛DNA重复序列(浮力密度1.709 g/cm3)0.005%(未加工)[29] SINE0.005%(加热)[30]猪、鸡和反刍动物SINE、LINE0.01%(加热)[31]反刍动物、马鹿、猪和牛/0.01%~0.10%(加热)[32]鸡肌动蛋白1%(加热)[33]猪
1.2 多重PCR-凝胶电泳法
多重PCR是设计多种普通PCR的引物,加入同一PCR反应体系里,同时扩增出多个核酸片段,以同时检测多种不同动物源成分的一项具有发展前景的技术[34]。相比于单一物种的PCR系统,多重PCR技术具有节省成本,提高分析速率、效率和可靠性的优点。表2汇总了运用多重PCR方法检测动物成分的相关研究。
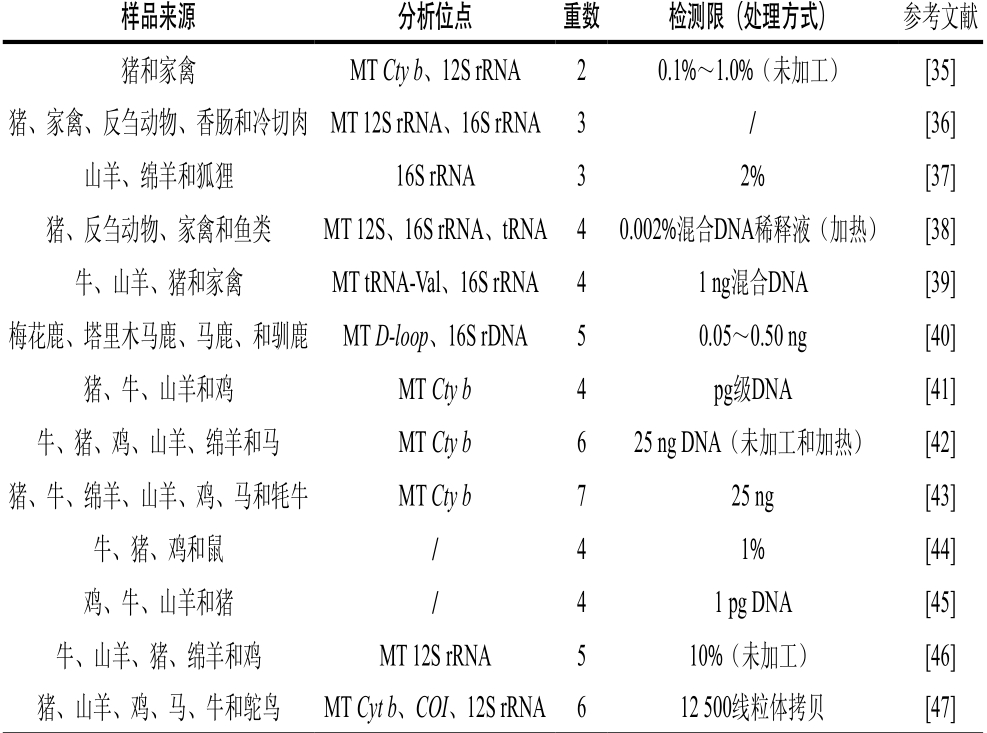
表2 多重PCR-凝胶电泳法的汇总表
Table 2 Reported multiplex PCR-gel electrophoresis methods
注:COI.细胞色素c氧化酶亚基Ⅰ(cytochrome c oxidase Ⅰ)基因。
样品来源分析位点重数检测限(处理方式)参考文献猪和家禽MT Cty b、12S rRNA20.1%~1.0%(未加工)[35]猪、家禽、反刍动物、香肠和冷切肉MT 12S rRNA、16S rRNA3/[36]山羊、绵羊和狐狸16S rRNA32%[37]猪、反刍动物、家禽和鱼类MT 12S、16S rRNA、tRNA40.002%混合DNA稀释液(加热)[38]牛、山羊、猪和家禽MT tRNA-Val、16S rRNA 41 ng混合DNA[39]梅花鹿、塔里木马鹿、马鹿、和驯鹿MT D-loop、16S rDNA50.05~0.50 ng[40]猪、牛、山羊和鸡MT Cty b4pg级DNA[41]牛、猪、鸡、山羊、绵羊和马MT Cty b625 ng DNA(未加工和加热)[42]猪、牛、绵羊、山羊、鸡、马和牦牛MT Cty b725 ng[43]牛、猪、鸡和鼠/41%[44]鸡、牛、山羊和猪/41 pg DNA[45]牛、山羊、猪、绵羊和鸡MT 12S rRNA510%(未加工)[46]猪、山羊、鸡、马、牛和鸵鸟MT Cyt b、COI、12S rRNA 612 500线粒体拷贝[47]
Matsunaga等[42]首次进行了多重PCR的研究,通过分析Cty b研究了牛、猪、山羊、鸡、绵羊、马肉的六重PCR,并得出了检测限是25 ng DNA。近几年,大量研究实现了在同一PCR体系里加入多对引物同时检测牛、猪、驴、山羊、绵羊、禽类等多种肉类成分的多重PCR鉴定。例如,Kitpipit等[47]用猪、鸡、马、牛、鸵鸟和山羊肉进行了六重PCR的研究,其分析目标是Cyt b、COI和12S rRNA基因,检测灵敏度可达到到12 500线粒体拷贝(相当于7 fg)。
1.3 PCR-RAPD法和PCR-RFLP法
食品中DNA成分复杂,加之PCR技术的高灵敏度,使得应用PCR扩增方法来鉴定近缘物种时具有因非特异性扩增而产生假阳性结果的缺点。因此,以PCR为基础,应用改良的引物设计策略或其他验证手段的鉴定方法为肉类定性鉴别提供了新方向。其中,PCR-RPLF与PCR-RAPD是两种主要策略。表3总结了部分至今为止有关RFLP法和RAPD法在肉类检测中的研究。
表3 PCR-RFLP和PCR-RAPD方法的汇总
Table 3 Reported PCR-RFLP and PCR-RAPD methods

技术物种分析位点检测限(处理方式)参考文献牛、猪、鸡和野猪M T D -l o o p 5 %(未加工)[ 4 8 ]牛、猪、鸡、反刍动物、火鸡、马、山羊和绵羊M T C t y b 0 . 5 %(加热)[ 4 9 ]牛、猪、鸡、山羊和绵羊M T C t y b 0 . 1 %(未加工)[ 5 0 ] P C R -R F L P山羊和鸭M T C t y b [ 5 1 ]牛、猪和山羊M T 1 2 S r R N A<1 %(加热)[ 5 2 ]牛M T C t y b / [ 5 3 ]牛和山羊/ / [ 5 4 ] P C R -R A P D猪、鸡、鸭、火鸡和鹅/ 2 5 0 p g稀释D N A [ 5 5 ] P C R -R A P D、P C R -R F L P牛、山羊、猪、鸡、反刍动物、火鸡、鸭和鸽子/ / [ 5 6 ]
PCR-RPLF法是将通用引物扩增后的PCR产物进行限制性内切酶酶切,然后通过凝胶电泳进行观察,作为单一物种的特有模式进行定性分析,可进行亲缘性较近的物种间的鉴别。RFLP分析技术用于区分动物成分,依赖于其特定的限制性酶切位点识别的差异。然而,PCR-RPLF的缺点是容易受到目标基因序列中酶切位点随机突变的影响,易产生不确定的检测结果,且RFLP方法绝大多数都是定性检测纯动物组织,因为混合物可能产生复杂的结果导致无法解释。
PCR-RAPD利用任意PCR短的引物扩增产生一系列的产物,得到PCR产物的指纹图谱,根据指纹图间的差别区分不同的种属。当参考材料可用的很少或DNA序列信息未知时,RAPD技术是非常强大的。但PCR-RAPD的重复性较差且受到食品中其他DNA成分的严重干扰,存在不易标准化与广泛应用的缺点。
1.4 DNA条形码
在生物物种鉴定领域,DNA条形码技术是发展最为迅速的一种新技术,最早由Hebert等[57]于2003年提出并用于物种鉴别和分类。目前DNA条形码技术的研究主要集中在鱼类[58-59]、动物[60]、植物[61]的成分鉴定。
表4 DNA条形码技术的汇总
Table 4 Reported DNA Barcoding methods
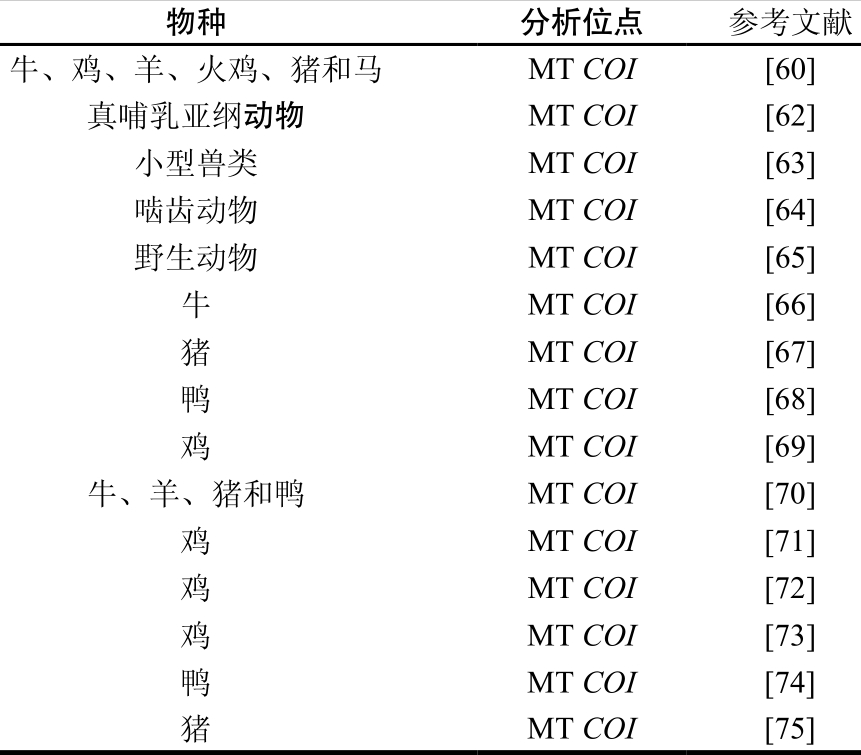
物种分析位点参考文献牛、鸡、羊、火鸡、猪和马MT COI[60]真哺乳亚纲动物MT COI[62]小型兽类MT COI[63]啮齿动物MT COI[64]野生动物MT COI[65]牛MT COI[66]猪MT COI[67]鸭MT COI[68]鸡MT COI[69]牛、羊、猪和鸭MT COI[70]鸡MT COI[71]鸡MT COI[72]鸡MT COI[73]鸭MT COI[74]猪MT COI[75]
DNA条形码技术最初用于生物分类和物种鉴别,后发展到应用于动物肉类鉴定中并得到广泛研究,是一种简单、快速、可靠、有效的分子鉴定技术。大多数动物物种的DNA条形码是线粒体上一段约650 bp编码基因COI,研究者将其作为动物鉴定的条形码标准片段[57]。表4总结了部分至今为止有关DNA条形码法在肉类检测中的研究。COI基因作为公认的DNA条形码被认为能够很好地对动物进行分类鉴定[74]。COI基因在双链环状闭合的核外线粒体基因组上,保证了其相对完整性和热稳定性,在深加工的肉类食品中有相对足够量的DNA被用于PCR扩增,确保足够高的PCR产物含量。COI序列还具有足够变异性、易扩增、片段自身在物种种内具有特异性和种间多样性等特点。然而,DNA条形码技术也存在局限性,首先,数据库中关于家禽家畜的DNA条形码序列很少,缺乏大量数据作为支撑[74]。其次,只适用于含有单一成分的鉴别,不能鉴别同一产品中的几种成分[75]。
虽然DNA条形码技术在动物成分鉴别中存在局限性,但DNA条形码技术能避免形态学分类的缺陷,对鉴定者的经验和专业知识背景要求较低,使科研和检疫检验工作更加高效,加速了物种鉴别进程。随着动物物种DNA条形码数据库的建立和不断完善,该技术将在肉类食品鉴别中发挥更完善的作用,为我国肉类食品安全检测和可追溯体系的建立提供参考。
1.5 实时荧光定量PCR技术
实时荧光定量PCR技术是利用荧光分子提供的荧光强度与PCR产物的丰度之间的相关性来实现实时数据采集[76]。相比于在末期进行分析的凝胶琼脂糖或聚丙烯酰胺电泳技术,实时荧光定量PCR方法自然更加精确灵敏。在复杂的混合而成的产品中,即使只有微量的不同物种成分,也能分析出来,因此被认为是在肉类鉴定中最有前途的分子工具之一[77]。研究人员利用实时荧光定量PCR技术对肉类成分定性定量检测进行了大量的研究,如表5汇总。
表5 实时荧光定量PCR法的汇总
Table 5 Reported real-time PCR methods

样品来源分析位点定量范围或误差(处理方式)参考文献牛MT ATP6、ATP80.001%[78]猪和牛MT DNA5% [79]牛MT Trna、ATP6、ATP80.001%(加热)[80]牛和猪PDEs、MY0.1%(未加工)[81]牛、猪和火鸡MT Cty b0.1%(未加工)[82]牛和猪MT tRNA ATP80.1%(加热)[83]牛GH0.02 ng[84]牛MY0.01% DNA稀释液(未加工)[84]反刍动物SINE10 fg稀释DNA(未加工)[85]牛、绵羊和山羊MT 18S rRNA0.1% DNA混合[86]狗和猫/4.0 pg DNA混合[87]猪、马和驴ND2、ND5、ATP 6、ATP 7、ATP 80.000 1 ng[88]猪/ 10 ng DNA[89]海鸥MT 12S rRNA、18S rRNA100 pg[90]牛、山羊、猪、马、鸡、火鸡和鹅/0.1% DNA混合物[91]鹿MY、LF2%~50%[92]半番鸭和鹅MT 12S rRNA1%(加热)[93]猪MT 12S rRNA0.5%(加热)[94]马和驴MT Cty b1 pg DNA液体(未加工)[95]山羊和绵羊MT 12S rRNA0.6%~10.0%[96]牛MT Cty b35 pg DNA (未加工)[97]黑线鳕TRF7% DNA(未加工)[98]反刍动物、牛、猪和鸡Bov-tA2、DNA重复序列(浮力密度1.711 g/cm3)、PRE-1、CR10.000 5% DNA混合物(未加工)[99]猪MT 12S rRNA、18S rRNA0.1% DNA混合[100]
续表5
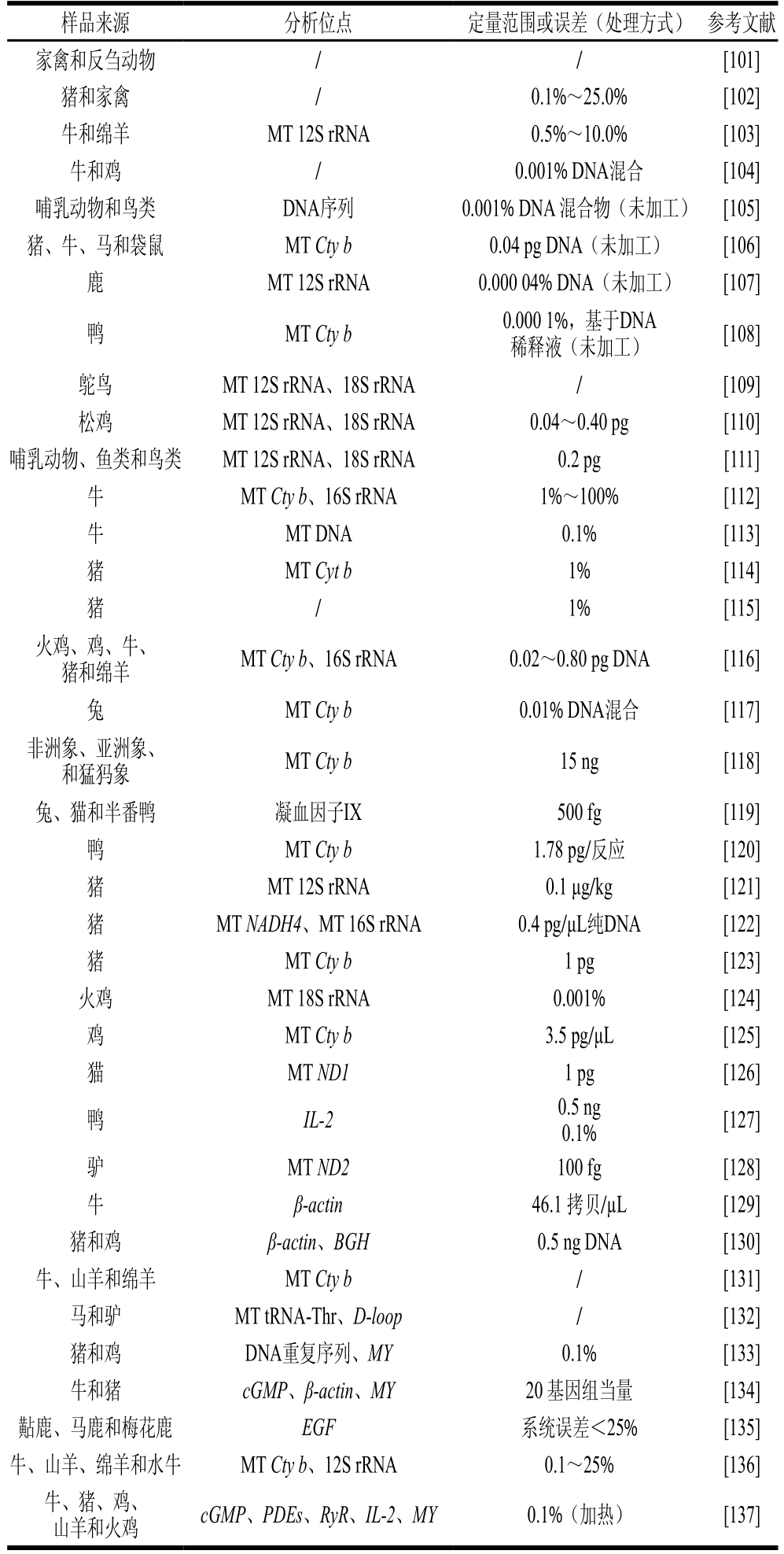
注:MY.肌肉生长抑制素(mmyostatin)基因;GH.生长激素(growth hormone)基因。
样品来源分析位点定量范围或误差(处理方式)参考文献家禽和反刍动物//[101]猪和家禽/0.1%~25.0%[102]牛和绵羊MT 12S rRNA0.5%~10.0%[103]牛和鸡/0.001% DNA混合[104]哺乳动物和鸟类DNA序列0.001% DNA 混合物(未加工)[105]猪、牛、马和袋鼠MT Cty b0.04 pg DNA(未加工)[106]鹿MT 12S rRNA0.000 04% DNA(未加工)[107]鸭MT Cty b0.000 1%,基于DNA稀释液(未加工)[108]鸵鸟MT 12S rRNA、18S rRNA/[109]松鸡MT 12S rRNA、18S rRNA0.04~0.40 pg [110]哺乳动物、鱼类和鸟类MT 12S rRNA、18S rRNA0.2 pg[111]牛MT Cty b、16S rRNA1%~100%[112]牛MT DNA0.1% [113]猪MT Cyt b1% [114]猪/ 1% [115]火鸡、鸡、牛、猪和绵羊MT Cty b、16S rRNA0.02~0.80 pg DNA[116]兔MT Cty b0.01% DNA混合[117]非洲象、亚洲象、和猛犸象MT Cty b15 ng[118]兔、猫和半番鸭凝血因子IX500 fg[119]鸭MT Cty b1.78 pg/反应[120]猪MT 12S rRNA0.1 μg/kg[121]猪MT NADH4、MT 16S rRNA0.4 pg/μL纯DNA[122]猪MT Cty b1 pg[123]火鸡MT 18S rRNA0.001%[124]鸡MT Cty b3.5 pg/μL[125]猫MT ND11 pg[126]鸭IL-20.5 ng 0.1% [127]驴MT ND2100 fg[128]牛β-actin46.1 拷贝/µL[129]猪和鸡β-actin、BGH0.5 ng DNA[130]牛、山羊和绵羊MT Cty b/[131]马和驴MT tRNA-Thr、D-loop/[132]猪和鸡DNA重复序列、MY0.1%[133]牛和猪 cGMP、β-actin、MY 20 基因组当量[134]黇鹿、马鹿和梅花鹿 EGF系统误差<25%[135]牛、山羊、绵羊和水牛MT Cty b、12S rRNA0.1~25% [136]牛、猪、鸡、山羊和火鸡cGMP、PDEs、RyR、IL-2、MY0.1%(加热)[137]
Lahiff等[78]在2002年首次利用荧光定量PCR(TaqMan探针法),通过分析线粒体tRNA ATP6和ATP8对热加工后牛肉成分检测进行了研究,检测限为0.001%。近年来肉类成分实时荧光PCR检测技术有了长足发展,但相关方法的完善与推广仍面临着挑战。肉类成分的定量分析通过已知样品中相同的目标基因(参照)绘制的浓度校准曲线与已知成分含量(m/m)进行对比分析建立。然而,在量化动物掺假时,动物组织组成、样品处理和肉类加工程度是不被研究者知道的,因此很难确定市场样品肉类含量(m/m)与测量方法的相关性。首先,实时定量PCR技术的准确度及定量性潜能受DNA产量的影响,比如由于加工处理的原因及同一样品不同组织中的DNA种类和生产细胞的数量存在有差异会影响DNA降解的程度。如经过粗加工和高度加工的肉类产品的定量结果相差约为10 倍[77]。其次,大多数动物细胞含有许多拷贝的线粒体DNA,并且在不同组织中的线粒体DNA拷贝数是不同的,导致未知样品中基于线粒体DNA序列测量结果和肉类含量(m/m)的相关性不准确。虽然该技术具有值得肯定的定量潜力,但很难实现肉类成分组织组成和加工过程尽可能与建立方法的条件保持一致,对商业肉制品中的目标物种进行定量测定仅仅只是可用,大部分研究主要还是利用实时荧光定量PCR的灵敏性对动物成分进行定性及半定量的判断。而且,由于使用了特殊的荧光探针,通常成本也相对较高。
可靠的荧光定量PCR检测方法必须考虑到DNA降解程度,才能推算出食品中原料肉的用量[137]。MY基因和GH基因被开发作为定量动物成分的校正参照标准。由于这些校正参照基因在不同哺乳动物及禽类组织中的表达水平相当,因此将对校正参照基因的定量结果与生肉组织中的定量结果相比,即可计算出基因组DNA的降解程度,从而进一步通过降解程度校正物种特异性扩增的定量结果。
1.6 ddPCR技术
ddPCR技术作为一种全新的准确定量核酸检测方法,通过把反应体系均分到大量反应单元中独立地进行PCR,并根据泊松分布和阳性比例来计算核酸数量。与传统PCR、定量PCR相比,其结果的精确度、准确性和灵敏度更佳。定量结果不再依赖于Ct值,直接给出靶序列的起始浓度,实现真正意义上的绝对定量。2014年Cai Yicun等[138]首次采用数字PCR技术,以DNA含量为中间值计算出DNA拷贝数与生鲜肉质量之间的线性关系,对肉制品中猪肉和鸡肉进行成分鉴定及含量的分析。王珊等[139]建立一种定量检测羊肉制品中羊源和猪源性成分的ddPCR方法,并将该方法与SN/T 2051—2008《食品、化妆品和饲料中牛羊猪源性成分检测方法 实时PCR法》中实时荧光定量PCR方法做对比,来检测3 份羊肉制品中的羊源和猪源性成分,得出结论:在肉种成分真伪鉴定上,ddPCR方法较实时荧光PCR方法更科学、准确。苗丽等[140]基于数字PCR技术,建立了定量检测肉及肉制品中牛肉和猪肉质量的方法。方法利用在一定范围内生鲜肉质量与DNA含量、DNA含量与DNA拷贝数之间均呈现明显的线性关系,以DNA含量为中间值计算出DNA拷贝数(C)与生鲜肉质量(M)之间的换算公式M牛= 0.062C-0.943、M猪=0.045C-1.72。对已知目标肉种含量的混合肉样进行检测,结果表明测量值和真实值基本一致,且不受外源物种的干扰。2015年Floren等[141]研究了基于ddPCR技术利用线粒体Cyt b基因151(马)、146(牛)、147 bp(猪)和染色体凝血因子Ⅱ基因(F2)95(马)、96 bp(牛)和97 bp(猪)作为标记基因鉴定和定量肉和肉制品。研究得出线粒体Cyt b基因作为标记基因进行物种量化是不合适的,利用染色体凝血因子Ⅱ基因(F2)作为标记基因,通过两步ddPCR,能实现牛、马、猪的精确量化,定量限(limit of quantity,LOQ)和检测限(limit of detection,LOD)分别在0.01%和0.001%。
ddPCR是一个拥有巨大潜力的新兴技术,具有高灵敏度、高精确度、高耐受性和绝对定量的优点,可以对诸如肉制品这种复杂样品中物种特异性靶基因实现更灵敏、更准确的检测,并且很容易将现成的实时荧光定量PCR检测体系进行直接转化,甚至无需优化。ddPCR技术将会在动物成分特别是精加工肉类产品的定量检测上进一步发展与完善,应用范围也会大大扩展。
2 基于核酸分子学方法的动物成分定量方法的优缺点
表6 动物成分物种鉴别定性、定量方法存在的优缺点
Table 6 Advantages and disadvantages of qualitative and quantitative PCR-based methods for species determination
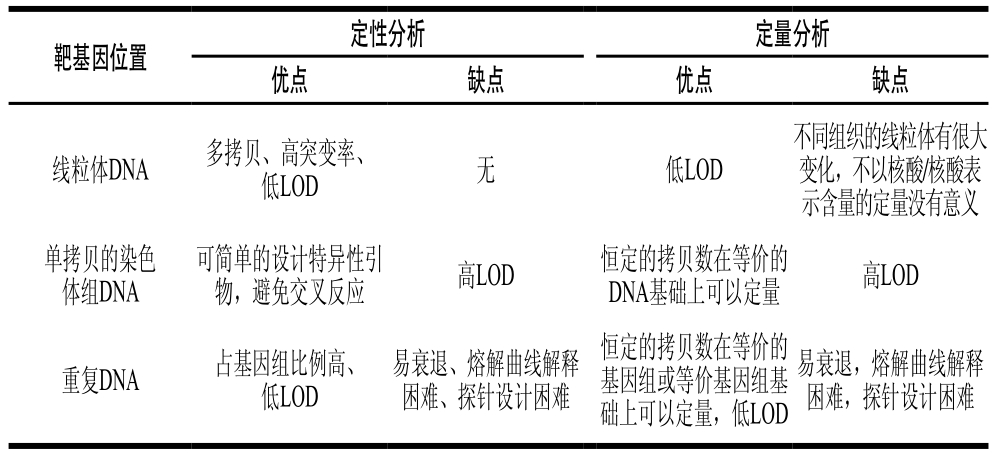
靶基因位置定性分析定量分析优点缺点优点缺点不同组织的线粒体有很大变化,不以核酸/核酸表示含量的定量没有意义单拷贝的染色体组DNA线粒体DNA多拷贝、高突变率、低LOD无低LOD可简单的设计特异性引物,避免交叉反应高LOD恒定的拷贝数在等价的DNA基础上可以定量高LOD重复DNA占基因组比例高、低LOD易衰退、熔解曲线解释困难、探针设计困难恒定的拷贝数在等价的基因组或等价基因组基础上可以定量,低LOD易衰退,熔解曲线解释困难,探针设计困难
基于核酸分子学水平的动物成分定性检测技术中一个重要的步骤是选择合适的靶基因。用于动物成分鉴别不同靶基因所存在的优缺点见表6。线粒体基因、染色体DNA和重复序列作为靶基因被广泛地用于识别野生和驯养肉类物种。如来源于线粒体的靶基因位点主要为线粒体Cty b、12S和16S核糖体RNA亚基、和D-loop等,这些都是动物源性成分检测最常用的标记基因。此外,核基因组特异性标记基因(生长激素基因、肌动蛋白基因、和黑毛发受体1基因)和重复序列(SINE、LINE)也可以有效鉴别肉类物种。在产品中检测到未标识的动物成分是相对简单的(掺假或不掺假),主要面临的挑战是定量的问题。在目标基因选择方面,线粒体基因组编码序列虽然是肉类定性鉴别的首选,但不同动物组织中线粒体数量区别较大,在动物组织种类未知的情况下以之作为定量检测的目标基因,则可能导致定量结果的偏差,而现有的部分研究却未考虑这一因素;细胞基因组中的重复序列不存在组织差异,但重复序列间的高同源性加大了特异性引物与探针设计的难度;细胞基因组中单拷贝的编码序列在食品加工过程中降解严重,导致检测灵敏度过低。
3 结 语
基于核酸DNA肉类成分定性分析的靶序列可来源于线粒体或基因组,包括单拷贝和重复序列,标记基因DNA序列的选择对检测方法的检测限有很大的影响。定量的分析应根据实时PCR或ddPCR分析,PCR扩增产物的序列必须来源于基因组,其中单拷贝和重复序列都可以使用。用线粒体DNA进行定量分析是不可能的,因为在未知样品中每个组织细胞的线粒体拷贝数是不确定的。
食品中肉类成分的种属鉴定技术是打击肉制品掺假、维护市场秩序的有效保障。如今,随着实时荧光定量PCR和ddPCR技术的飞速发展为肉类成分检测开辟了新的途径,使得食品中肉类成分的定量分析与溯源成为可能。在定量检测中,通过对反应体系的精巧设计而提升方法的准确性与实用价值将成为相关技术的发展方向与趋势。
参考文献:
[1] MONTOWSKA M, POSPIECH E. Species identification of meat by electrophoretic methods[J]. Acta Scientiarum Polonorum Technologia Alimentaria, 2007, 6(1): 5-16.
[2] CHOU C C, LIN S P, LEE K M, et al. Fast differentiation of meats from fifteen animal species by liquid chromatography with electrochemical detection using copper nanoparticle plated electrodes[J]. Journal of Chromatography B: Biomedical Sciences and Applications, 2007, 846(1/2): 230-239. DOI:10.1016/ j.jchromb.2006.09.006.
[3] ELLIS D I, BROADHURST D, CLARKE S J, et al. Rapid identification of closely related muscle foods by vibrational spectroscopy and machine learning[J]. Analyst, 2005, 130(12): 1648-1654. DOI:10.1039/b511484e.
[4] BALLIN N Z, VOGENSEN F K, KARLSSON A H. Species determination-can we detect and quantify meat adulteration?[J]. Meat Science, 2009, 83(2): 165-174. DOI:10.1016/j.meatsci.2009.06.003.
[5] TARTAGLIA M, SAULLE E, PESTALOZZA S, et al. Detection of bovine mitochondrial DNA in ruminant feeds: a molecular approach to test for the presence of bovine-derived materials[J]. Journal of Food Protection, 1998, 61(5): 513-518. DOI:10.1128/JB.00923-09.
[6] WANG R F, MYERS M J, CAMPBELL W, et al. A rapid method for PCR detection of bovine materials in animal feedstuffs[J]. Molecular and Cellular Probes, 2000, 14(1): 1-5. DOI:10.1006/mcpr.1999.0273.
[7] COLGAN S, O’BRIEN L, MAHER M, et al. Development of a DNA-based assay for species identification in meat and bone meal[J]. Food Research International, 2001, 34(5): 409-414. DOI:0.1016/S0963-9969(00)00185-X.
[8] MYERS M J, FRIEDMAN S L, FARRELL D E, et al. Validation of a polymerase chain reaction method for the detection of rendered bovine-derived materials in feedstuffs[J]. Journal of Food Protection, 2001, 64(4): 564-566.
[9] LAHIFF S, GLENNON M, BRIEN L, et al. Species-specific PCR for the identification of ovine, porcine and chicken species in meat and bone meal (MBM)[J]. Molecular and Cellular Probes, 2001, 15(1): 27-35. DOI:10.1006/mcpr.2000.0336.
[10] KRCMAR P, RENCOVA E. Identification of species-specific DNA in feedstuffs[J]. Journal of Agricultural and Food Chemistry, 2004, 51(26): 7655-7658. DOI:10.1021/jf034167y.
[11] KUSAMA T, NOMURA T, KADOWAKI K. Development of primers for detection of meat and bone meal in ruminant feed and identification of the animal of origin[J]. Journal of Food Protection, 2004, 67(6): 1289-1292.
[12] BELLAGAMBA F, VALFRE F, PANSERI S, et al. Polymerase chain reaction-based analysis to detect terrestrial animal protein in fish meal[J]. Journal of Food Protection, 2003, 66(4): 682-685.
[13] HA J C, JUNG W T, NAM Y S, et al. PCR identification of ruminant tissue in raw and heat-treated meat meals[J]. Journal of Food Protection, 2006, 69(9): 2241-2247.
[14] MARTIN I, GARCIA T, FAJARDO V, et al. Species-specific PCR for the identification of ruminant species in feedstuffs[J]. Meat Science, 2007, 75(1): 120-127. DOI:10.1016/j.meatsci.2006.06.019.
[15] RODR☒GUEZ M A, GARC☒A T, GONZ☒LEZ I, et al. Identification of goose, mule duck, chicken, turkey, and swine in foie gras by speciesspecific polymerase chain reaction[J]. Journal of Agricultural and Food Chemistry, 2003, 51(6): 1524-1529. DOI:10.1021/jf025784+.
[16] YIN R H, BAI W L, WANG J M, et al. Development of an assay for rapid identification of meat from yak and cattle using polymerase chain reaction technique[J]. Meat Science, 2009, 83(1): 38-44.
[17] SAKARIDIS I, GANOPOULOS I, ARGIRIOU A, et al. A fast and accurate method for controlling the correct labeling of products containing buffalo meat using high resolution melting (HRM) analysis[J]. Meat Science, 2013, 94(1): 84-88. DOI:10.1016/ j.meatsci.2012.12.017.
[18] CHAUMPLUK P, CHIKAE M, TAKAMURA Y, et al. Novel electrochemical identification and semi quantification of bovine constituents in feedstuffs[J]. Science and Technology of Advanced Materials, 2006, 7(3): 263-269. DOI:10.1016/j.stam.2006.03.001.
[19] 蒋立, 胡茂. 种属特异性P C R技术在检测鹅肥肝掺假中的应用研究[J]. 畜禽业, 2010(8): 54-55. DOI:10.3969/ j.issn.1008-0414.2010.08.031.
[20] KORTBAOUI R, LOCAS A, IMBEAU M, et al. Universal mitochondrial PCR combined with species-specific dot-blot assay as a source-tracking method of human, bovine, chicken, ovine, and porcine in fecal-contaminated surface water[J]. Water Research, 2009, 43(7): 2002-2010. DOI:10.1016/j.watres.2009.01.030.
[21] MANE B G, MENDIRATTA S K, TIWARI A K. Beef specific polymerase chain reaction assay for authentication of meat and meat products[J]. Food Control, 2012, 28(2): 246-249. DOI:10.1016/ j.foodcont.2012.05.031.
[22] PASCOAL A, PRADO M, CALO P, et al. Detection of bovine DNA in raw and heat-processed foodstuffs, commercial foods and specific risk materials by a novel specific polymerase chain reaction method[J]. European Food Research and Technology, 2005, 220(3/4): 444-450. DOI:10.1007/s00217-004-1088-x.
[23] 王思伟, 刘静静, 李芸, 等. 山羊线粒体cytB基因片段特异性PCR反应条件优化[J]. 中国草食动物科学, 2014(增刊 1): 92-94. DOI:10.3969/j.issn.2095-3887.2014.z1.035.
[24] 李通, 尹艳, 王海, 等. 聚合酶链式反应快速鉴别5 种常见肉类别[J].食品科学, 2013, 34(8): 249-252. DOI:10.7506/spkx1002-6630-201308054.
[25] MANE B G, MENDIRATTA S K, TIWARI A K. Polymerase chain reaction assay for identification of chicken in meat and meat products[J]. Food Chemistry, 2009, 116(3): 806-810. DOI:10.1016/ j.foodchem.2009.03.030.
[26] KARABASANAVARA N S, SINGHA S P, UMAPATHI V, et al. A highly specific PCR assay for identification of raw and heat treated mutton (Ovis aries)[J]. Small Ruminant Research, 2011, 100(2/3): 153-158. DOI:10.1016/j.smallrumres.2011.07.009.
[27] 李通, 尹艳, 袁其朋, 等. 运用PCR方法鉴别四种犬科动物的研究[J]. 食品工业科技, 2013, 34(17): 146-149. DOI:10.13386/ j.issn1002-0306.2013.17.075.
[28] ZHANG G L, ZHENG M G, ZHOU Z J, et al. Establishment and application of a polymerase chain reaction for the identification of beef[J]. Meat Science, 1999, 51(3): 233-236.
[29] CALVO J H, RODELLAR C, ZARAGOZA P, et al. Beef- and bovinederived material identification in processed and unprocessed food and feed by PCR amplification[J]. Journal of Agricultural and Food Chemistry, 2002, 50(19): 5262-5264. DOI:10.1021/jf020051a.
[30] CALVO J H, ZARAGOZA P, OSTA R. Technical note: a quick and more sensitive method to identify pork in processed and unprocessed food by PCR amplification of a new specific DNA fragment[J]. Journal of Animal Science, 2001, 79(8): 2108-2112.
[31] TAJIMA K, ENISHI O, AMARI M, et al. PCR detection of DNAs of animal origin in feed by primers based on sequences of short and long interspersed repetitive elements[J]. Bioscience, Biotechnology, and Biochemistry, 2002, 66(10): 2247-2250. DOI:10.1271/bbb.66.2247.
[32] AMARAL J S, SANTOS C G, MELO V S, et al. Authentication of a traditional game meat sausage (Alheira) by species-specific PCR assays to detect hare, rabbit, red deer, pork and cow meats[J]. Food Research International, 2014, 60(6): 140-145. DOI:10.1016/j.foodres.2013.11.003.
[33] HOPWOOD A J, FAIRBROTHER K S, LOCKLEY A K, et al. An actin gene-related polymerase chain reaction (PCR) test for identification of chicken in meat mixtures[J]. Meat Science, 1999, 53(4): 227-231. DOI:10.1016/S0309-1740(99)00060-1.
[34] TOBE S S, LINACRE A M T. A multiplex assay to identify 18 European mammal species from mixtures using the mitochondrial cytochrome b gene[J]. Electrophoresis, 2008, 29(2): 340-347.
[35] SOARES S, AMARAL J S, MAFRA I, et al. Quantitative detection of poultry meat adulteration with pork by a duplex PCR assay[J]. Meat Science, 2010, 85(3): 531-536. DOI:10.1016/j.meatsci.2010.03.001.
[36] GHOVVATI S, NASSIRI M R, MIRHOSEINI S Z. Fraud identification in industrial meat products by multiplex PCR assay[J]. Food Control, 2009, 20(8): 696-699. DOI:10.1016/j.foodcont.2008.09.002.
[37] 张全芳, 马德源, 刘艳艳, 等. 利用多重PCR技术检测羊肉中掺杂狐狸肉的方法研究[J]. 山东农业科学, 2014(12): 4-6. DOI:10.3969/ j.issn.1001-4942.2014.12.002.
[38] DALMASSO A, FONTANELLA E, PIATTI P, et al. A multiplex PCR assay for the identification of animal species in feedstuffs[J]. Molecular and Cellular Probes, 2004, 18(2): 81-87. DOI:10.1016/ j.mcp.2003.09.006.
[39] ZHA D M, XING X M, YANG F H. A multiplex PCR assay for fraud identification of deer products[J]. Food Control, 2010, 21(10): 1402-1407.
[40] ZHA D M, XING X M, YANG F H. Rapid identification of deer products by multiplex PCR assay[J]. Food Chemistry, 2011, 129(4): 1904-1908.
[41] 何玮玲, 张驰, 杨静, 等. 食品中4 种肉类成分多重PCR的快速鉴别方法[J]. 中国农业科学, 2012, 45(9): 1873-1880. DOI:10.3864/ j.issn.0578-1752.2012.09.024.
[42] MATSUNAGA T, CHIKUNI K, TANABE R, et al. A quick and simple method for the identification of meat species and meat products by PCR assay[J]. Meat Science, 1999, 51(2): 143-148. DOI:10.1016/ S0309-1740(98)00112-0.
[43] 冯海永, 韩建林. 羊肉产品中若干动物源性成分的七重PCR检测技术应用研究[J]. 中国畜牧兽医, 2010, 37(9): 85-90.
[44] 苏葳艺, 李欣南, 于雷, 等. 利用多重PCR方法检测牛肉中的掺假肉[J].食品工业, 2015(2): 277-280.
[45] ZHANG C. Semi-nested multiplex PCR enhanced method sensitivity of species detection in further-processed meats[J]. Food Control, 2013, 31(2): 326-330. DOI:10.1016/j.foodcont.2012.11.002.
[46] RASTOGI G, DHARNE M, BHARDE A, et al. Species determination and authentication of meat samples by mitochondrial 12S rRNA gene sequence analysis and conformation-sensitive gel electrophoresis[J]. Current Science, 2004, 87(9): 1278-1281.
[47] KITPIPIT T, SITTICHAN K, THANAKIATKRAI P. Direct-multiplex PCR assay for meat species identification in food products[J]. Food Chemistry, 2014, 163(3): 77-82. DOI:10.1016/j.foodchem.2014.04.062.
[48] MONTIEL-SOSA J F, RUIZ-PESINI E, MONTOYA J, et al. Direct and highly species-specific detection of pork meat and fat in meat products by PCR amplification of mitochondrial DNA[J]. Journal of Agricultural and Food Chemistry, 2000, 48(7): 2829-2832. DOI:10.1021/jf9907438.
[49] BELLAGAMBA F, MORETTI V M, COMINCINI S, et al. Identification of species in animal feedstuffs by polymerase chain reaction-restriction fragment length polymorphism analysis of mitochondrial DNA[J]. Journal of Agricultural and Food Chemistry, 2001, 49(8): 3775-3781. DOI:10.1021/jf0010329.
[50] MAEDE D. A strategy for molecular species detection in meat and meat products by PCR-RFLP and DNA sequencing using mitochondrial and chromosomal genetic sequences[J]. European Food Research and Technology, 2006, 224(2): 209-217. DOI:10.1007/ s00217-006-0320-2.
[51] 冯海永, 刘丑生, 何建文, 等. 利用线粒体DNA Cyt b基因PCR-RFLP分析方法鉴别羊肉和鸭肉[J]. 食品工业科技, 2012, 33(13): 319-321. DOI:10.13386/j.issn1002-0306.2012.13.093.
[52] SUN Y L, LIN C S. Establishment and application of a fluorescent polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method for identifying porcine, caprine, and bovine meats[J]. Journal of Agricultural and Food Chemistry, 2003, 51(7): 1771-1776. DOI:10.1021/jf020860u.
[53] 高琳, 徐幸莲, 周光宏. 肉制品中牛源性成分的PCR-RFLP检测[J]. 食品科技, 2007, 32(4): 191-193. DOI:10.3969/j.issn.1005-9989.2007.04.057.
[54] 田金辉, 李宝明, 尉婷媛, 等. T-RFLP快速鉴定生鲜肉中牛羊成分的研究[J]. 中国畜牧兽医, 2011, 38(3): 84-86.
[55] CALVO J H, ZARAGOZA P, OSTA R. Random amplified polymorphic DNA fingerprints for identification of species in poultry pate[J]. Poultry Science, 2001, 80(4): 522-524. DOI:10.1093/ ps/80.4.522.
[56] HAUNSHI S, BASUMATARY R, GIRISH P S, et al. Identification of chicken, duck, pigeon and pig meat by species-specific markers of mitochondrial origin[J]. Meat Science, 2009, 83(3): 454-459. DOI:10.1016/j.meatsci.2009.06.026.
[57] HEBERT P D N, RATNASINGHAM S, de WAARD J R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species[J]. Proceedings of the Royal Society B: Biological Science, 2003, 270(Suppl 1): 96-99. DOI:10.1098/rsbl.2003.0025.
[58] 李新光, 王璐, 赵峰, 等. DNA条形码技术在鱼肉及其制品鉴别中的应用[J]. 食品科学, 2013, 34(18): 337-342. DOI:10.7506/spkx1002-6630-201318069.
[59] 王敏, 刘荭, 黄海, 等. DNA条形码技术在深圳鱼肉制品鉴定中的应用[J]. 食品科学, 2015, 36(20): 247-251. DOI:10.7506/spkx1002-6630-201520048.
[60] KANE D E, HELLBERG R S R. Identification of species in ground meat products sold on the U.S. commercial market using DNA-based methods[J]. Food Control, 2016, 59: 158-163. DOI:10.1016/ j.foodcont.2015.05.020.
[61] CBOL Plant Working Group. A DNA barcode for land plants[J]. Proceedings of the National Academy of Sciences of the USA, 2009, 106(31): 12794-12797. DOI:10.1073/pnas.0905845106.
[62] LUO A R, ZHANG A B, HO S Y W, et al. Potential efficacy of mitochondrial genes for animal DNA barcoding: a case study using eutherian mammals[J]. BMC Genomics, 2011, 12(1): 1-13.
[63] 马英, 李海龙, 鲁亮, 等. DNA条形码技术在青海海东地区小型兽类鉴定中的应用[J]. 生物多样性, 2012, 20(2): 193-198.
[64] M☒LLER L, GONCALVES G L, CORDEIRO-ESTRELA P, et al. DNA barcoding of sigmodontine rodents: identifying wildlife reservoirs of zoonoses[J]. PLoS ONE, 2013, 8(11): e80282.
[65] QUINTO C A, TINOCO R, HELLBERG R S R. DNA barcoding reveals mislabeling of game meat species on the US commercial market[J]. Food Control, 2016, 59: 386-392. DOI:10.1016/ j.foodcont.2015.05.043.
[66] CAI Y S, ZHANG L, SHEN F J, et al. DNA barcoding of 18 species of Bovidae[J]. Chinese Science Bulletin, 2011, 56(2): 164-168.
[67] 欧阳解秀, 王立贤. DNA条形码技术在地方猪种质资源保护中的应用[J]. 农业生物技术学报, 2013, 21(3): 348-354. DOI:10.3969/ j.issn.1674-7968.2013.03.012.
[68] 徐向明. 我国3 个地方品种鸭线粒体DNACOⅠ基因的DNA条形码初步分析[J]. 畜牧与兽医, 2008, 40(11): 51-53.
[69] 高玉时, 屠云洁, 童海兵, 等. 6 个地方鸡种线粒体COⅠ基因的DNA条形码[J]. 农业生物技术学报, 2007, 15(6): 924-930. DOI:10.3969/ j.issn.1674-7968.2007.06.003.
[70] 王爽, 李永波, 马超峰, 等. DNA条形码COⅠ序列在常见肉类鉴别中的应用研究[J]. 现代食品科技, 2016, 32(1): 188-193. DOI:10.13982/j.mfst.1673-9078.2016.1.030.
[71] 高玉时, 唐修君, 屠云洁, 等. 基于线粒体COⅠ基因15 个鸡种的DNA编码研究[J]. 中国农业科学, 2011, 44(3): 587-594. DOI:10.3864/j.ssn.0578-1752.2011.03.020.
[72] 唐修君, 高玉时, 屠云洁, 等. 基于线粒体COⅠ基因的2 个新发现鸡种资源DNA编码研究[J]. 中国畜牧, 2011, 38(1): 133-136.
[73] 屠云洁, 陈国宏, 高玉时, 等. 3 个地方鸡种线粒体DNA COⅠ基因条形码遗传多样性研究[J]. 家畜生态学报, 2009, 30(1): 16-19. DOI:10.3969/j.issn.1673-1182.2009.01.004.
[74] 吕冬梅, 黄原, 文慧, 等. DNA条形码技术在食品鉴定中的应用[J].食品科学, 2015, 36(9): 248-253. DOI:10.7506/spkx1002-6630-201509046.
[75] HELLBERG R S R, MORRISSEY M T. Advances in DNA-based techniques for the detection of seafood species substitution on the commercial market[J]. Journal of Laboratory Automation, 2011, 16(4): 308-321. DOI:0.1016/j.jala.2010.07.004.
[76] LOPEZ-ANDREO M, LUGO L, GARRIDO-PERTIERRA A, et al. Identification and quantitation of species in complex DNA mixtures by real-time polymerase chain reaction[J]. Analytical Biochemistry, 2005, 339(1): 73-82. DOI:10.1016/j.ab.2004.11.045.
[77] KOPPEL R, ZIMMERLI F, BREITENMOSER A. Heptaplex real-time PCR for the identification and quantification of DNA from beef, pork, chicken, turkey, horse meat, sheep (mutton) and goat[J]. European Food Research and Technology, 2009, 230(1): 125-133.
[78] LAHIFF S, GLENNON M, LYNG J, et al. Real-time polymerase chain reaction detection of bovine DNA in meat and bone meal samples[J]. Journal of Food Protection, 2002, 65(7): 1158-1165.
[79] LPEZ-ANDREO M, ALDEGUER M, GUILL☒N I, et al. Detection and quantification of meat species by qPCR in heat-processed food containing highly fragmented DNA[J]. Food Chemistry, 2012, 134(1): 518-523. DOI:10.1016/j.foodchem.2012.02.111.
[80] LAUBE I, SPIEGELBERG A, BUTSCHKE A, et al. Methods for the detection of beef and pork in foods using real-time polymerase chain reaction[J]. International Journal of Food Science and Technology, 2003, 38(2): 111-118. DOI:10.1046/j.1365-2621.2003.00651.x.
[81] DOOLEY J J, PAINE K E, GARRET S D, et al. Detection of meat species using TaqMan real-time PCR assays[J]. Meat Science, 2004, 68(3): 431-438. DOI:10.1016/j.meatsci.2004.04.010.
[82] FUMIERE O, DUBOIS M, BAETEN V, et al. Effective PCR detection of animal species in highly processed animal byproducts and compound feeds[J]. Analytical and Bioanalytical Chemistry, 2006, 385(6): 1045-1054. DOI:10.1007/s00216-006-0533-z.
[83] INES L, JUTTA Z, ALMUTH S, et al. Development and design of a‘ready-to-use’ reaction plate for a PCR-based simultaneous detection of animal species used in foods[J]. International Journal of Food Science and Technology, 2006, 42(1): 9-17.
[84] BRODMANN P D, MOOR D. Sensitive and semi-quantitative TaqManTMreal-time polymerase chain reaction systems for the detection of beef (Bos taurus) and the detection of the family Mammalia in food and feed[J]. Meat Science, 2003, 65(1): 599-607. DOI:10.1016/S0309-1740(02)00253-X.
[85] MENDOZA-ROMERO L, VERKAAR E L, SAVELKOUL P H, et al. Real-time PCR detection of ruminant DNA[J]. Journal of Food Protection, 2004, 67(3): 550-554.
[86] PEGELS N, GONZ☒LEZ I, MART☒N I, et al. Applicability assessment of a real-time PCR assay for the specific detection of bovine, ovine and caprine material in feedstuffs[J]. Food Control, 2011, 22(8): 1189-1196. DOI:10.1016/j.foodcont.2011.01.015.
[87] KANTHASWAMY S, PREMASUTHAN A. Quantitative realtime PCR (qPCR) assay for human-dog-cat species identification and nuclear DNA quantification[J]. Forensic Science International Genetics, 2012, 6(2): 290-295. DOI:10.1016/j.fsigen.2011.06.005.
[88] KESMEN Z, GULLUCE A, SAHIN F, et al. Identification of meat species by TaqMan-based real-time PCR assay[J]. Meat Science, 2009, 82(4): 444-449. DOI:10.1016/j.meatsci.2009.02.019.
[89] ALI M E, HASHIM U, MUSTAFA S, et al. Analysis of pork adulteration in commercial meatballs targeting porcine-specific mitochondrial cytochrome b gene by TaqMan probe real-time polymerase chain reaction[J]. Meat Science, 2012, 91(4): 454-459. DOI:10.1016/j.meatsci.2012.02.031.
[90] KESMEN Z, CELEBI Y, G☒LL☒CE A, et al. Detection of seagull meat in meat mixtures using real-time PCR analysis[J]. Food Control, 2013, 34(1): 47-49. DOI:10.1016/j.foodcont.2013.04.006.
[91] CREMONESI P, PISANI L F, LECCHI C, et al. Development of 23 individual TaqMan☒real-time PCR assays for identifying common foodborne pathogens using a single set of amplification conditions[J]. Food Microbiology, 2014, 43: 35-40.
[92] DRUML B, MAYER W, CICHNA-MARKL M, et al. Development and validation of a TaqMan real-time PCR assay for the identification and quantification of roe deer (Capreolus capreolus) in food to detect food adulteration[J]. Food Chemistry, 2015, 178: 319-326. DOI:10.1016/j.foodchem.2015.01.003.
[93] RODR☒GUEZ M A, GARC☒A T, GONZ☒LEZ I, et al. Quantitation of mule duck in goose foie gras using TaqMan real-time polymerase chain reaction[J]. Journal of Agricultural and Food Chemistry, 2004, 52(6): 1478-1483. DOI:10.1021/jf035240n.
[94] RODR☒GUEZ M A, GARC☒A T, GONZ☒LEZ I, et al. TaqMan real-time PCR for the detection and quantitation of pork in meat mixtures[J]. Meat Science, 2005, 70(1): 113-120. DOI:10.1016/ j.meatsci.2004.12.005.
[95] CHRISHOLM J, CONYERS C, BOOTH C, et al. The detection of horse and donkey using real-time PCR[J]. Meat Science, 2005, 70(4): 727-732. DOI:10.1016/j.meatsci.2005.03.009.
[96] LPEZ-CALLEJA I, GONZ☒LEZ I, FAJARDO V, et al. Quantitative detection of goats’ milk in sheep’s milk by real-time PCR[J]. Food Control, 2007, 18(11): 1466-1473.
[97] ZHANG C L, FOWLER M R, SCOTT N W, et al. A TaqMan real-time PCR system for the identification and quantification of bovine DNA in meats, milks and cheeses[J]. Food Control, 2007, 18(9): 1149-1158. DOI:10.1016/j.foodcont.2006.07.018.
[98] HIRD H J, HOLD G L, CHISHOLM J, et al. Development of a method for the quantification of haddock (Melanogrammus aeglefinus) in commercial products using real-time PCR[J]. European Food Research and Technology, 2005, 220(5): 633-637. DOI:10.1007/ s00217-004-1050-y.
[99] WALKER J A, HUGHES D A, ANDERS B A, et al. Quantitative intra-short interspersed element PCR for species-specific DNA identification[J]. Analytical Biochemistry, 2003, 316(2): 259-269. DOI:10.1016/S0003-2697(03)00095-2.
[100] MART☒N I, GARC☒A T, FAJARDO V, et al. SYBR-Green realtime PCR approach for the detection and quantification of pig DNA in feedstuffs[J]. Meat Science, 2009, 82(2): 252-259. DOI:10.1016/ j.meatsci.2009.01.023.
[101] SAKALAR E, ABASIYANIK M F. The devolopment of duplex realtime PCR based on SYBR Green florescence for rapid identification of ruminant and poultry origins in foodstuff[J]. Food Chemistry, 2012, 130(4): 1050-1054.
[102] SOARES S, AMARAL J S, OLIVEIRA M B P P. A SYBR Green real-time PCR assay to detect and quantify pork meat in processed poultry meat products[J]. Meat Science, 2013, 94(1): 115-120. DOI:10.1016/j.meatsci.2012.12.012.
[103] LPEZ-CALLEJA I, GONZ☒LEZ I, FAJARDO V, et al. Real-time TaqMan PCR for quantitative detection of cows’ milk in ewes’ milk mixtures[J]. International Dairy Journal, 2007, 17(7): 729-736.
[104] SAFDAR M, JUNEJO Y. Development and validation of fast duplex real-time PCR assays based on SYBER Green florescence for detection of bovine and poultry origins in feedstuffs[J]. Food Chemistry, 2015, 173: 660-664. DOI:10.1016/j.foodchem.2014.10.088.
[105] WALKER J A, HUGHES D A, HEDGES D J, et al. Quantitative PCR for DNA identification based on genome-specific interspersed repetitive elements[J]. Genomics, 2004, 83(3): 518-527. DOI:10.1016/ j.ygeno.2003.09.003.
[106] LPEZANDREO M, GARRIDOPERTIERRA A A, PUYET A. Evaluation of postpolymerase chain reaction melting temperature analysis for meat species identification in mixed DNA samples[J]. Journal of Agricultural and Food Chemistry, 2006, 54(21): 7973-7978.
[107] FAJARDO V, GONZ☒LEZ I, MART☒N I, et al. Real-time PCR for detection and quantification of red deer (Cervus elaphus), fallow deer (Dama dama), and roe deer (Capreolus capreolus) in meat mixtures[J]. Meat Science, 2008, 79(2): 289-298. DOI:10.1016/ j.meatsci.2007.09.013.
[108] HIRD H, CHISHOLM J, BROWN J. The detection of commercial duck species in food using a single probe-multiple species-specific primer real-time PCR assay[J]. European Food Research and Technology, 2005, 221(3): 559-563. DOI:10.1007/s00217-005-1197-1.
[109] ROJAS M, GONZ☒LEZ I, PAV☒N M ☒, et al. Application of a real-time PCR assay for the detection of ostrich (Struthio camelus) mislabelling in meat products from the retail market[J]. Food Control, 2011, 22(3/4): 523-531. DOI:10.1016/j.foodcont.2010.09.039.
[110] ROJAS M, GONZ☒LEZ I, PAV☒N M ☒, et al. Development of a real-time PCR assay to control the illegal trade of meat from protected capercaillie species (Tetrao urogallus)[J]. Forensic Science International, 2011, 210(1/2/3): 133-138. DOI:10.1016/ j.forsciint.2011.02.021.
[111] BENEDETTO A, ABETE M C, SQUADRONE S. Towards a quantitative application of real-time PCR technique for fish DNA detection in feedstuffs[J]. Food Chemistry, 2011, 126(3): 1436-1442. DOI:10.1016/j.foodchem.2010.11.131.
[112] DRUMMOND M G, BRASIL B S A F, DALSECCO L S, et al. A versatile real-time PCR method to quantify bovine contamination in buffalo products[J]. Food Control, 2013, 29(1): 131-137. DOI:10.1016/ j.foodcont.2012.05.051.
[113] SAWYER J, WOOD C, SHANAHAN D, et al. Real-time PCR for quantitative meat species testing[J]. Food Control, 2003, 14(8): 579-583. DOI:10.1016/S0956-7135(02)00148-2.
[114] DEMIRHAN Y, ULCA P, SENYUVA H Z. Detection of porcine DNA in gelatine and gelatine-containing processed food products-Halal/Kosher authentication[J]. Meat Science, 2012, 90(3): 686-689. DOI:10.1016/j.meatsci.2011.10.014.
[115] ABDULMAWJOOD A, KRISCHEK C, WICKE M, et al. Determination of pig sex in meat and meat products using multiplex real time-PCR[J]. Meat Science, 2012, 91(3): 272-276. DOI:10.1016/ j.meatsci.2012.02.001.
[116] CAMM☒C, DOMENICO M D, MONACO F. Development and validation of fast Real-Time PCR assays for species identification in raw and cooked meat mixtures[J]. Food Control, 2012, 23(2): 400-404.
[117] SANTOS C G, MELO V S, AMARAL J S, et al. Identification of hare meat by a species-specific marker of mitochondrial origin[J]. Meat Science, 2012, 90(3): 836-841. DOI:10.1016/j.meatsci.2011.10.018.
[118] KO Y H, KIM M S, BANG J, et al. Real-time PCR detection and quantification of elephantid DNA: species identification for highly processed samples associated with the ivory trade[J]. Forensic Science International, 2012, 219(1/2/3): 106-112.
[119] CHANG J T, CHEN Y C, CHOU Y C, et al. Quantitative detection of residual porcine host cell DNA by real-time PCR[J]. Biologicals Journal of the International Association of Biological Standardization, 2014, 42(2): 74-78. DOI:10.1016/j.biologicals.2013.10.005.
[120] 张驰, 邱皓璞, 张筠. 荧光定量PCR检测肉制品中鸭源性成分[J].食品科学, 2013, 34(18): 154-157. DOI:10.7506/spkx1002-6630-201318031.
[121] 王颖, 史艳宇, 刘金华, 等. 荧光定量PCR方法检测畜肉食品中猪源性成分[J]. 食品安全质量检测学报, 2013(5): 1529-1534.
[122] 周彤, 李家鹏, 田寒友, 等. 一种基于实时荧光聚合酶链式反应的肉及肉制品中猪源性成分含量测定[J]. 肉类研究, 2013, 27(12): 11-15.
[123] 范丽丽, 李培, 傅春玲, 等. 实时荧光聚合酶链式反应法检测食品中猪源性成分[J]. 食品科学, 2013, 34(8): 224-227. DOI:10.7506/ spkx1002-6630-201308048.
[124] 张舒亚, 谌鸿超, 宋青, 等. 食品和饲料中火鸡源性成分的实时荧光PCR检测方法[J]. 食品与生物技术学报, 2013, 32(2): 207-211. DOI:10.3969/j.issn.1673-1689.2013.02.016.
[125] 范丽丽, 李培, 傅春玲, 等. 食品中鸡源性成分实时荧光PCR检测方法的建立[J]. 食品科学, 2014, 35(2): 248-251. DOI:10.7506/ spkx1002-6630-201402048.
[126] 高晓丽, 杨昕霆, 王丹, 等. 实时荧光聚合酶链式反应法检测食品中猫源性成分[J]. 食品科学, 2014, 35(24): 235-238. DOI:10.7506/ spkx1002-6630-201424045.
[127] 程欣, 何玮玲, 黄明. 实时荧光PCR法检测食品中鸭肉成分[J]. 食品科学, 2013, 34(24): 92-96. DOI:10.7506/spkx1002-6630-201324019.
[128] 高晓丽, 杨昕霆, 薛晨玉, 等. 实时荧光聚合酶链式反应法检测食品中狗源性成分[J]. 食品工业科技, 2015, 36(2): 61-64. DOI:10.13386/ j.issn1002-0306.2015.02.004.
[129] 苗丽, 李志娟, 王珊, 等. 肉制品中牛源性成分荧光定量聚合酶链式反应方法的建立与应用[J]. 肉类研究, 2015, 29(9): 30-33. DOI:10.15922/j.cnki.rlyj.2015.09.007.
[130] 何玮玲, 胡序建, 程欣, 等. 含有扩增内标的食品中猪肉和鸡肉成分Taqman探针实时荧光PCR检测方法的建立[J]. 中国农业科学, 2013, 46(21): 4578-4585. DOI:10.3864/j.issn.0578-1752.2013.21.021.
[131] 曾少灵, 秦智锋, 阮周曦, 等. 多重实时荧光PCR检测牛、山羊和绵羊源性成分[J]. 生物工程学报, 2009, 25(1): 139-146. DOI:10.3321/ j.issn:1000-3061.2009.01.021.
[132] 吴亚君, 王斌, 刘鸣畅, 等. 阿胶中马和驴成分的实时荧光PCR检测[J]. 食品科学, 2014, 35(8): 85-88. DOI:10.7506/spkx1002-6630-201408016.
[133] BALLIN N Z, VOGENSEN F K, KARLSSON A H. PCR amplification of repetitive sequences as a possible approach in relative species quantification[J]. Meat Science, 2011, 90(2): 438-443. DOI:10.1016/j.meatsci.2011.09.002.
[134] IWOBI A, SEBAH D, KRAEMER I, et al. A multiplex real-time PCR method for the quantification of beef and pork fractions in minced meat[J]. Food Chemistry, 2015, 169: 305-313. DOI:10.1016/ j.foodchem.2014.07.139.
[135] DRUML B, GRANDITS S, MAYER W, et al. Authenticity control of game meat products-A single method to detect and quantify adulteration of fallow deer (Dama dama), red deer (Cervus elaphus) and sika deer (Cervus nippon) by real-time PCR[J]. Food Chemistry, 2015, 170: 508-517. DOI:10.1016/j.foodchem.2014.08.048.
[136] INES L, JUTTA Z, HERMANN B. Quantitative determination of commercially relevant species in foods by real-time PCR[J]. International Journal of Food Science and Technology, 2007, 42(3): 336-341.
[137] BALLIN N Z, VOGENSEN F K, KARLSSON A H. Species determination - Can we detect and quantify meat adulteration?[J]. Meat Science, 2009, 83(2): 165-174. DOI:10.1016/j.meatsci.2009.06.003.
[138] CAI Yicun, LI Xiang, L☒ Rong, et al. Quantitative analysis of pork and chicken products by droplet digital PCR[J]. Biomed Research International, 2014: 1-6. DOI:10.1155/2014/810209.
[139] 王珊, 李志娟, 苗丽. 微滴式数字PCR与实时荧光PCR检测羊肉制品中羊源和猪源性成分方法的比较[J]. 肉类工业, 2015(7): 38-41. DOI:10.3969/j.issn.1008-5467.2015.07.012.
[140] 苗丽, 张秀平, 陈静, 等. 数字PCR法对肉制品中牛源和猪源成分的定量分析[J]. 食品科学, 2016, 37(8): 187-191. DOI:10.7506/ spkx1002-6630-201608033.
[141] FLOREN C, WIEDEMANN I, BRENIG B, et al. Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR)[J]. Food Chemistry, 2014, 173: 1054-1058. DOI:10.1016/j.foodchem.2014.10.138.
A Review of Current DNA-Based Methodologies for Meat Authentication
WANG Jinbin1,2,3, LI Wen1,3,*, BAI Lan1,3, LIU Hua1,3, JIANG Wei1,3, WU Xiao1,3, WANG Rongtan4, TANG Xueming1,2,3,*
(1. Shanghai Academy of Agricultural Sciences, Shanghai 201106, China; 2. College of Food Science and Technology, Shanghai Ocean University, Shanghai 200090, China; 3. Key Laboratory of Agricultural Genetics and Breeding, Shanghai 201106, China; 4. Shanghai Ruifeng Agricultural Sci-Tech Company Ltd., Shanghai 201106, China)
Abstract:In recent years, the problem of meat adulteration has occurred frequently. Full attention has been paid to DNA-based methodologies for meat species identi☒cation because of their high sensitivity and speci☒city, as well as rapid processing time and low cost. This article presents an overview of the commonly used DNA-based methodologies to verify the authenticity of meat and meat products with focus on their detection limits. Moreover, this review highlights the current applications and future prospects of real-time fluorescence quantitative polymerase chain reaction (PCR) and digital PCR in the identification of animal origin ingredients. Finally, target genes from different sources (nuclear DNA and mitochondrial DNA) are compared in terms of their characteristics and their in☒uence on the sensitivity and speci☒city of species identi☒cation and quanti☒cation.
Key words:adulteration; meat products; digital PCR; species determination
DOI:10.7506/spkx1002-6630-201711049
中图分类号:TS207.3
文献标志码:A
文章编号:1002-6630(2017)11-0318-10引文格式:
收稿日期:2016-05-12
基金项目:上海市闵行区产学研合作计划项目(2016MH256);上海市农委青年人才成长计划项目(沪农青字(2014)第1-20号)作者简介:王金斌(1982—),男,助理研究员,博士研究生,研究方向为食品安全与检测技术。E-mail:wangjinbin2013@126.com
*通信作者:李文(1982—),女,助理研究员,硕士,研究方向为功能食品技术研发和利用。E-mail:liwen@saas.sh.cn唐雪明(1970—),男,研究员,博士,研究方向为生物技术。E-mail:xueming70@foxmail.com






